{
"validmind.data_validation.DatasetDescription": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {}
},
"validmind.data_validation.ClassImbalance": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {
"min_percent_threshold": 10
}
},
"validmind.data_validation.Duplicates": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {
"min_threshold": 1
}
},
"validmind.data_validation.HighCardinality": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {
"num_threshold": 100,
"percent_threshold": 0.1,
"threshold_type": "percent"
}
},
"validmind.data_validation.MissingValues": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {
"min_threshold": 1
}
},
"validmind.data_validation.Skewness": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {
"max_threshold": 1
}
},
"validmind.data_validation.UniqueRows": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {
"min_percent_threshold": 1
}
},
"validmind.data_validation.TooManyZeroValues": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {
"max_percent_threshold": 0.03
}
},
"validmind.data_validation.IQROutliersTable": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {
"threshold": 1.5
}
},
"validmind.data_validation.IQROutliersBarPlot": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {
"threshold": 1.5,
"fig_width": 800
}
},
"validmind.data_validation.DescriptiveStatistics": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {}
},
"validmind.data_validation.PearsonCorrelationMatrix": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {}
},
"validmind.data_validation.HighPearsonCorrelation": {
"inputs": {
"dataset": "raw_dataset"
},
"params": {
"max_threshold": 0.3,
"top_n_correlations": 10,
"feature_columns": null
}
},
"validmind.model_validation.ModelMetadata": {
"inputs": {
"model": "model"
},
"params": {}
},
"validmind.data_validation.DatasetSplit": {
"inputs": {
"datasets": [
"train_dataset",
"test_dataset"
]
},
"params": {}
},
"validmind.model_validation.sklearn.PopulationStabilityIndex": {
"inputs": {
"datasets": [
"train_dataset",
"test_dataset"
],
"model": "model"
},
"params": {
"num_bins": 10,
"mode": "fixed"
}
},
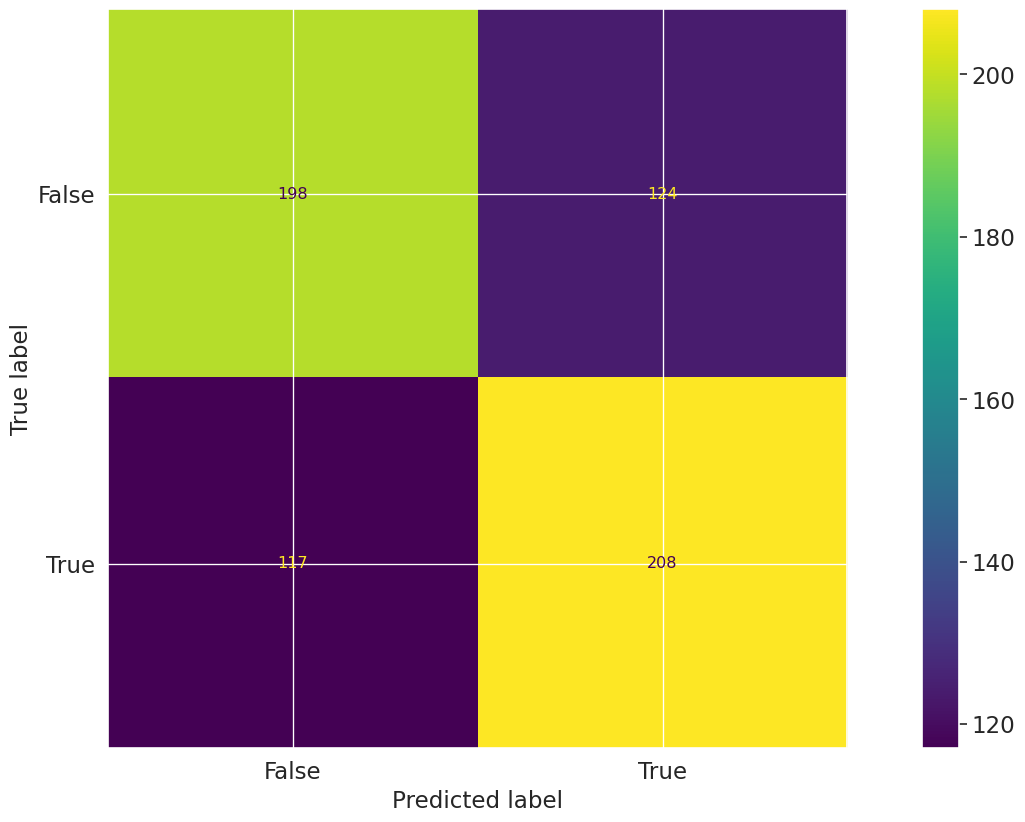
"validmind.model_validation.sklearn.ConfusionMatrix": {
"inputs": {
"dataset": "test_dataset",
"model": "model"
},
"params": {
"threshold": 0.5
}
},
"validmind.model_validation.sklearn.ClassifierPerformance:in_sample": {
"inputs": {
"model": "model",
"dataset": "train_dataset"
}
},
"validmind.model_validation.sklearn.ClassifierPerformance:out_of_sample": {
"inputs": {
"model": "model",
"dataset": "test_dataset"
}
},
"validmind.model_validation.sklearn.PrecisionRecallCurve": {
"inputs": {
"model": "model",
"dataset": "test_dataset"
},
"params": {}
},
"validmind.model_validation.sklearn.ROCCurve": {
"inputs": {
"model": "model",
"dataset": "test_dataset"
},
"params": {}
},
"validmind.model_validation.sklearn.TrainingTestDegradation": {
"inputs": {
"datasets": [
"train_dataset",
"test_dataset"
],
"model": "model"
},
"params": {
"max_threshold": 0.1
}
},
"validmind.model_validation.sklearn.MinimumAccuracy": {
"inputs": {
"dataset": "test_dataset",
"model": "model"
},
"params": {
"min_threshold": 0.7
}
},
"validmind.model_validation.sklearn.MinimumF1Score": {
"inputs": {
"dataset": "test_dataset",
"model": "model"
},
"params": {
"min_threshold": 0.5
}
},
"validmind.model_validation.sklearn.MinimumROCAUCScore": {
"inputs": {
"dataset": "test_dataset",
"model": "model"
},
"params": {
"min_threshold": 0.5
}
},
"validmind.model_validation.sklearn.PermutationFeatureImportance": {
"inputs": {
"model": "model",
"dataset": "test_dataset"
},
"params": {
"fontsize": null,
"figure_height": null
}
},
"validmind.model_validation.sklearn.SHAPGlobalImportance": {
"inputs": {
"model": "model",
"dataset": "test_dataset"
},
"params": {
"kernel_explainer_samples": 10,
"tree_or_linear_explainer_samples": 200,
"class_of_interest": null
}
},
"validmind.model_validation.sklearn.WeakspotsDiagnosis": {
"inputs": {
"datasets": [
"train_dataset",
"test_dataset"
],
"model": "model"
},
"params": {
"features_columns": null,
"metrics": null,
"thresholds": null
}
},
"validmind.model_validation.sklearn.OverfitDiagnosis": {
"inputs": {
"model": "model",
"datasets": [
"train_dataset",
"test_dataset"
]
},
"params": {
"metric": null,
"cut_off_threshold": 0.04
}
},
"validmind.model_validation.sklearn.RobustnessDiagnosis": {
"inputs": {
"datasets": [
"train_dataset",
"test_dataset"
],
"model": "model"
},
"params": {
"metric": null,
"scaling_factor_std_dev_list": [
0.1,
0.2,
0.3,
0.4,
0.5
],
"performance_decay_threshold": 0.05
}
}
}