| validmind.data_validation.ClassImbalance |
Class Imbalance |
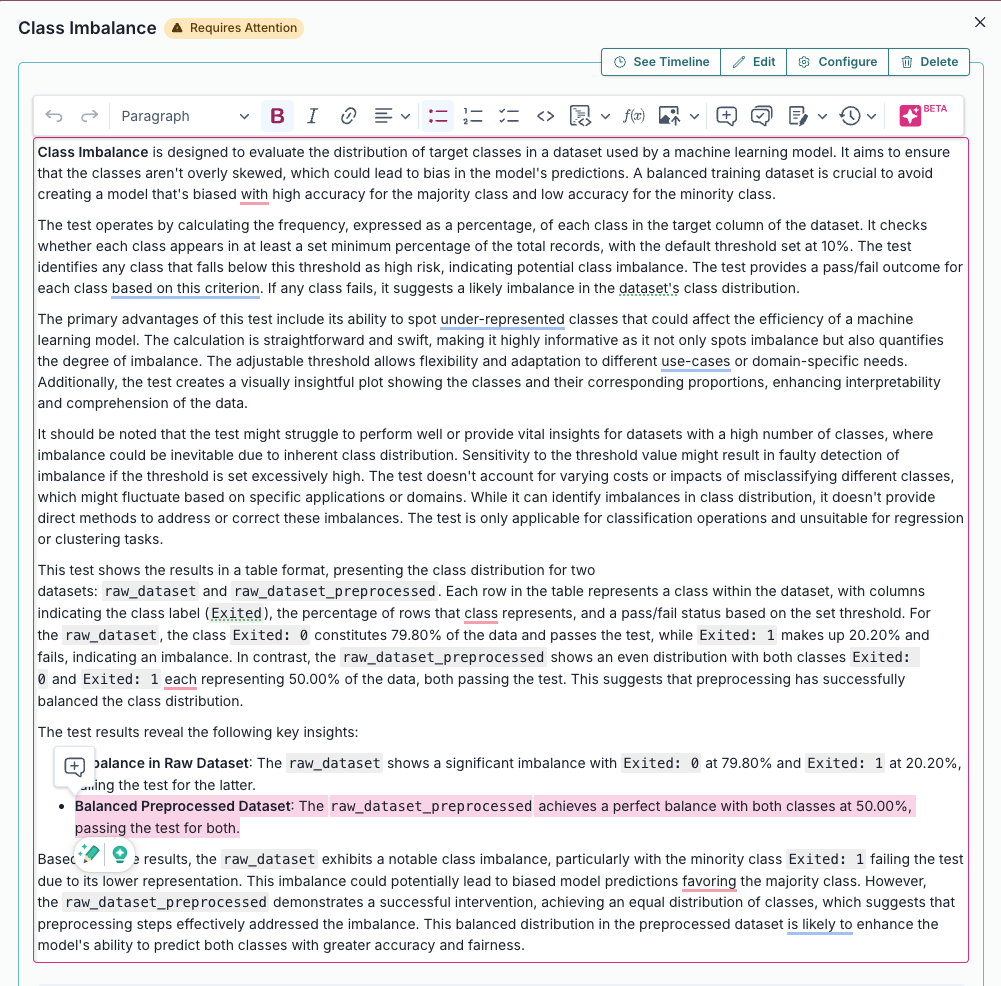
Evaluates and quantifies class distribution imbalance in a dataset used by a machine learning model.... |
True |
True |
['dataset'] |
{'min_percent_threshold': {'type': 'int', 'default': 10}} |
['tabular_data', 'binary_classification', 'multiclass_classification', 'data_quality'] |
['classification'] |
| validmind.data_validation.DescriptiveStatistics |
Descriptive Statistics |
Performs a detailed descriptive statistical analysis of both numerical and categorical data within a model's... |
False |
True |
['dataset'] |
{} |
['tabular_data', 'time_series_data', 'data_quality'] |
['classification', 'regression'] |
| validmind.data_validation.Duplicates |
Duplicates |
Tests dataset for duplicate entries, ensuring model reliability via data quality verification.... |
False |
True |
['dataset'] |
{'min_threshold': {'type': '_empty', 'default': 1}} |
['tabular_data', 'data_quality', 'text_data'] |
['classification', 'regression'] |
| validmind.data_validation.HighCardinality |
High Cardinality |
Assesses the number of unique values in categorical columns to detect high cardinality and potential overfitting.... |
False |
True |
['dataset'] |
{'num_threshold': {'type': 'int', 'default': 100}, 'percent_threshold': {'type': 'float', 'default': 0.1}, 'threshold_type': {'type': 'str', 'default': 'percent'}} |
['tabular_data', 'data_quality', 'categorical_data'] |
['classification', 'regression'] |
| validmind.data_validation.HighPearsonCorrelation |
High Pearson Correlation |
Identifies highly correlated feature pairs in a dataset suggesting feature redundancy or multicollinearity.... |
False |
True |
['dataset'] |
{'max_threshold': {'type': 'float', 'default': 0.3}, 'top_n_correlations': {'type': 'int', 'default': 10}, 'feature_columns': {'type': 'list', 'default': None}} |
['tabular_data', 'data_quality', 'correlation'] |
['classification', 'regression'] |
| validmind.data_validation.MissingValues |
Missing Values |
Evaluates dataset quality by ensuring missing value ratio across all features does not exceed a set threshold.... |
False |
True |
['dataset'] |
{'min_threshold': {'type': 'int', 'default': 1}} |
['tabular_data', 'data_quality'] |
['classification', 'regression'] |
| validmind.data_validation.MissingValuesBarPlot |
Missing Values Bar Plot |
Assesses the percentage and distribution of missing values in the dataset via a bar plot, with emphasis on... |
True |
False |
['dataset'] |
{'threshold': {'type': 'int', 'default': 80}, 'fig_height': {'type': 'int', 'default': 600}} |
['tabular_data', 'data_quality', 'visualization'] |
['classification', 'regression'] |
| validmind.data_validation.Skewness |
Skewness |
Evaluates the skewness of numerical data in a dataset to check against a defined threshold, aiming to ensure data... |
False |
True |
['dataset'] |
{'max_threshold': {'type': '_empty', 'default': 1}} |
['data_quality', 'tabular_data'] |
['classification', 'regression'] |
| validmind.plots.BoxPlot |
Box Plot |
Generates customizable box plots for numerical features in a dataset with optional grouping using Plotly.... |
True |
False |
['dataset'] |
{'columns': {'type': 'Optional', 'default': None}, 'group_by': {'type': 'Optional', 'default': None}, 'width': {'type': 'int', 'default': 1800}, 'height': {'type': 'int', 'default': 1200}, 'colors': {'type': 'Optional', 'default': None}, 'show_outliers': {'type': 'bool', 'default': True}, 'title_prefix': {'type': 'str', 'default': 'Box Plot of'}} |
['tabular_data', 'visualization', 'data_quality'] |
['classification', 'regression', 'clustering'] |
| validmind.plots.HistogramPlot |
Histogram Plot |
Generates customizable histogram plots for numerical features in a dataset using Plotly.... |
True |
False |
['dataset'] |
{'columns': {'type': 'Optional', 'default': None}, 'bins': {'type': 'Union', 'default': 30}, 'color': {'type': 'str', 'default': 'steelblue'}, 'opacity': {'type': 'float', 'default': 0.7}, 'show_kde': {'type': 'bool', 'default': True}, 'normalize': {'type': 'bool', 'default': False}, 'log_scale': {'type': 'bool', 'default': False}, 'title_prefix': {'type': 'str', 'default': 'Histogram of'}, 'width': {'type': 'int', 'default': 1200}, 'height': {'type': 'int', 'default': 800}, 'n_cols': {'type': 'int', 'default': 2}, 'vertical_spacing': {'type': 'float', 'default': 0.15}, 'horizontal_spacing': {'type': 'float', 'default': 0.1}} |
['tabular_data', 'visualization', 'data_quality'] |
['classification', 'regression', 'clustering'] |
| validmind.stats.DescriptiveStats |
Descriptive Stats |
Provides comprehensive descriptive statistics for numerical features in a dataset.... |
False |
True |
['dataset'] |
{'columns': {'type': 'Optional', 'default': None}, 'include_advanced': {'type': 'bool', 'default': True}, 'confidence_level': {'type': 'float', 'default': 0.95}} |
['tabular_data', 'statistics', 'data_quality'] |
['classification', 'regression', 'clustering'] |